DLRC Achieves Cyber Essentials Plus Certification
Published Apr 11, 2025
Published 01st July 2024

Drug-drug interactions (DDI) are a common problem during drug treatment and give rise to a large number of hospital admissions as a result of medically important, sometimes serious or even fatal adverse events. Drug-drug interactions can also cause partial or complete abolishment of treatment efficacy. Several drugs have been withdrawn from the market as a result of drug-drug interactions that were only discovered post-marketing. The potential for drug-drug interactions is considered in the benefit-risk evaluation of a medicinal product. It can negatively impact this balance either through increased incidence of adverse events or reduced efficacy.
The results of DDI evaluations inform the protocols for clinical studies in patients regarding the use of concomitant drugs. Information about the interaction potential should be gained as early in drug development as practically possible to assure safety and avoid unnecessary restrictions of concomitant medications and/or exclusion of patients who require the concomitant medications in clinical studies, typically phase 2/3 studies.
The ICH M12 guideline recommends a consistent approach to designing, conducting, and interpreting enzyme- or transporter-mediated in vitro and clinical DDI studies during the development of an investigational drug. This article provides an overview of ICH M12 guideline focusing on small molecules and in vitro DDI and highlight some considerations for your DDI Gap analysis.
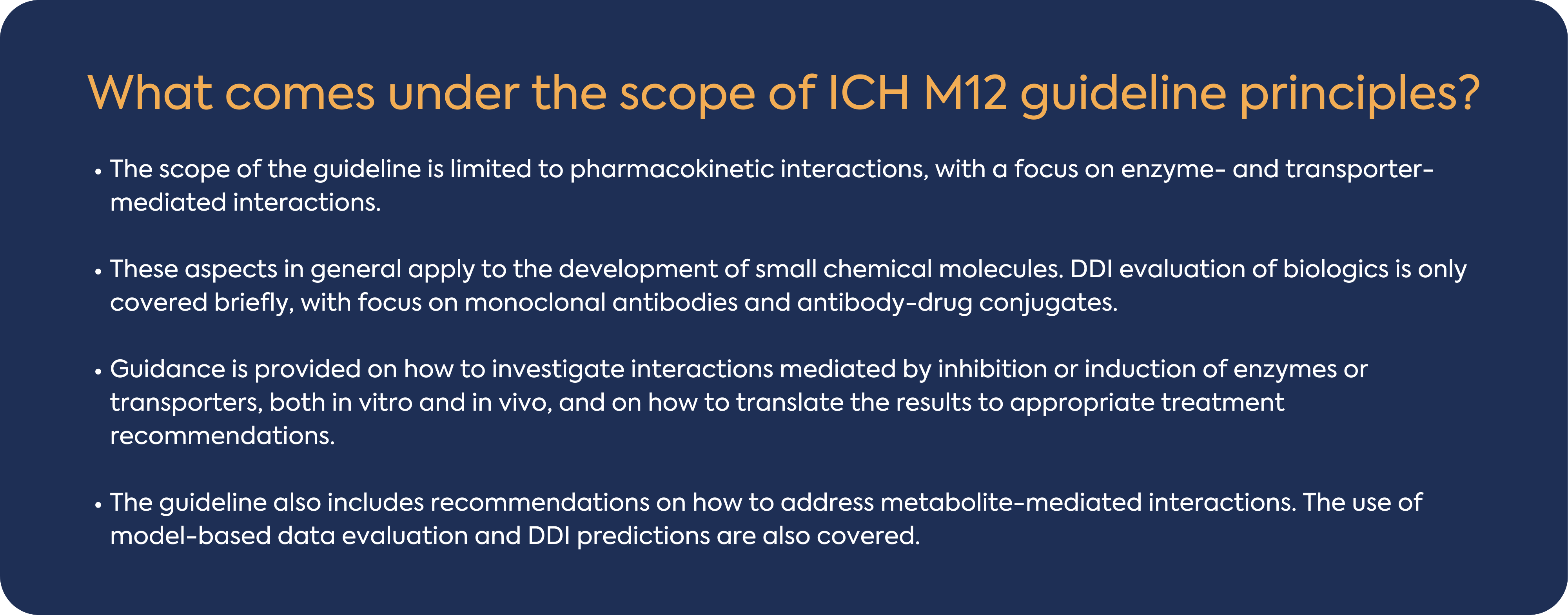
In general, a drug that is metabolized by multiple enzymes has a lower DDI potential than a drug that is metabolized by a single enzyme. Multiple metabolic pathways reduce the likelihood of DDIs as the closure of one metabolic pathway (e.g., by concomitant medicines) is unlikely to result in complete loss of the elimination pathway. Enzymes involved in metabolic pathways which contribute t ≥ 25% of drug elimination should be identified. The guideline indicates that CYP1A2, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, and CYP3A should be routinely evaluated using in vitro reaction phenotyping experiments to determine which enzymes metabolize the investigational drug, typically this work is performed early in development. If the investigational drug is not found to undergo significant in vitro metabolism by these major CYP enzymes, then additional assessments may be needed to understand which enzymes contribute to the overall metabolism.
Cytochrome P450 enzymes metabolize a significant number of small molecule drugs on the market today and, as such, are a major focus of DDI assessments. Inhibition of CYP450 is a major cause of PK-based DDIs, and drugs that are inhibitors of CYP450 enzymes reduce the clearance of other drugs that are metabolized by inhibited enzymes. Based on the ICH M12 guideline, both direct and time-dependent inhibition should be tested for DDI risk assessment for all major CYP enzymes e.g., CYP1A2, CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2D6, and CYP3A. The assay should be conducted in pooled HLM, pooled hepatocytes, microsomes from recombinant systems and should include the determination of the Ki (or the IC50) using several concentrations of the investigational drug relevant to the clinical setting.
The ICH M12 guideline indicated that considering the generally limited magnitude of UGT inhibition-mediated DDIs, a routine evaluation of investigational drugs to inhibit UGTs may not be warranted. If direct glucuronidation is one of the major elimination pathways of an investigational drug, it is recommended to evaluate in vitro whether the drug can inhibit UGTs including UGT1A1 and UGT2B7. When an investigational drug is to be used with another drug that is mainly metabolized by direct glucuronidation, it is recommended to evaluate the in vitro potential inhibitory effect of the investigational drug on the UGT isoform(s) responsible for the elimination of the other drug.
Enzyme induction can lead to decreased efficacy and/or increased formation of toxic metabolites. The guideline recommends testing for CYP1A2, CYP2B6 and CYP3A4 induction, the first instance, as markers of induction mediated via PXR/CAR (CYP3A4, CYP2B6) and AhR (CYP1A2). If no induction of CYP3A4 enzymes is observed, evaluating the induction potential of CYP2C enzymes is not necessary because both CYP3A4 and CYP2C enzymes are induced via activation of the pregnane X receptor (PXR). These assays should be conducted in human hepatocytes from a minimum 3 individual donors and recommended readout (with the exception of CYP2C19) is changes in CYP450 mRNA levels.
In vitro P-gp substrate and inhibition data are typically expected in regulatory submissions. Initial in vitro assessments are used to predict DDI potential, aiding the development of a P-gp transporter-based clinical drug interaction strategy. In addition, regulatory agencies consider BCRP, a clinically important drug transporter, and an in vitro assessment of both substrate and inhibitory potency is expected for registration.
OATP1B1 and OATP1B3 are relevant to the hepatic uptake and disposition of drugs, and drug interactions.
OAT1, OAT3, and OCT2 are renal transporters expressed on the basolateral membrane of the renal proximal tubule. MATE1 and MATE2-K are expressed on the brush border membrane. All the aforementioned renal transporters can play a role in the active renal secretion of investigational drugs. OAT1 and 3 constitute the first step in the active renal tubular secretion of negatively charged drugs and as such are potentially implicated in renal DDIs and toxicity.
All test systems and experimental conditions for the assessment of transporther mediated DDIs should be validated and the acceptance criteria for study results should be established.
Based on the current guideline, in vitro methods to evaluate transporter induction are not well established. If an investigational drug has been observed to be an inducer of CYP enzymes via activation of nuclear receptors such as PXR or CAR, it is likely that transporters regulated through these receptors will be induced, such as P-gp.
The risk of a DDI when the metabolite acts as a substrate should be evaluated for a pharmacologically active metabolite if: it contributes to in vivo target effect to a similar or greater extent than the parent drug. If plasma protein binding of the parent drug and the metabolite is high, it is preferred to determine their protein binding in the same study to reduce inter-study variability.
An in vitro CYP enzyme inhibition study should be conducted if: the AUC (metabolite) ≥ 25% of AUC (parent), and also account for at least 10% of drug-related material in circulation (i.e., considered as major metabolite often determined based on radioactivity data).
When the drug is a prodrug or a metabolite is mainly formed extra‑hepatically, in vitro evaluation of a metabolite’s induction potential on CYP enzymes is recommended if the metabolite is a major metabolite and has AUC (metabolite) / AUC (parent) ≥ 25%.

ICH M12 guideline provides general recommendations on how to assess the DDI potential of investigational drugs. It is recognised that DDI assessments are often tailored to the specific drug, target patient population, and therapeutic context. Alternative methods can be used if they meet the requirements of the applicable statutes and regulations. The focus of this guideline is on the development of new drugs, but additional DDI evaluation should be considered if new scientific information about DDI potential becomes available after drug approval.
Please contact the DLRC at hello@dlrcgroup.com with any questions you may have – our experienced team of non-clinical regulatory experts are here to assist you in the advancement of your drug interaction studies and will participate in these discussions with you to facilitate your communication with the regulatory agencies.

Published Apr 11, 2025

Published Mar 27, 2025

Published Mar 06, 2025
Published Feb 26, 2025

Published Feb 25, 2025

Published Feb 03, 2025

Published Feb 03, 2025

Published Jan 30, 2025

Published Jan 30, 2025